gmx_MMPBSA Calculations¶
Here we describe a series of frequent issues related to calculations and their possible solutions.
Note
Most of the errors noted here are the result of inconsistent input files. Please read the documentation and make sure your files are consistent.
ValueError: could not convert string to float: '*************'
-
This error has two possible causes:
-
The structure defined in
-cs,-rs, or-lsoptions is inconsistent, or the trajectory has not been fitted (remove PBC) properly. This is the most common error. Many times it is because the system under study is longer than some edges of the box.Possible solutions:¶
Check for structure consistency
Visualize the structure contained in the structure input file given in the
-cs,-rs, or-lsoptions and make sure it is consistent (as shown in Fig 1, right panel). On the other hand, if the structure is "broken" (as shown in Fig 1, left panel) this could generate inconsistent results.Generate the structure from tpr file
gmx editconf -f md.tpr -o md.pdb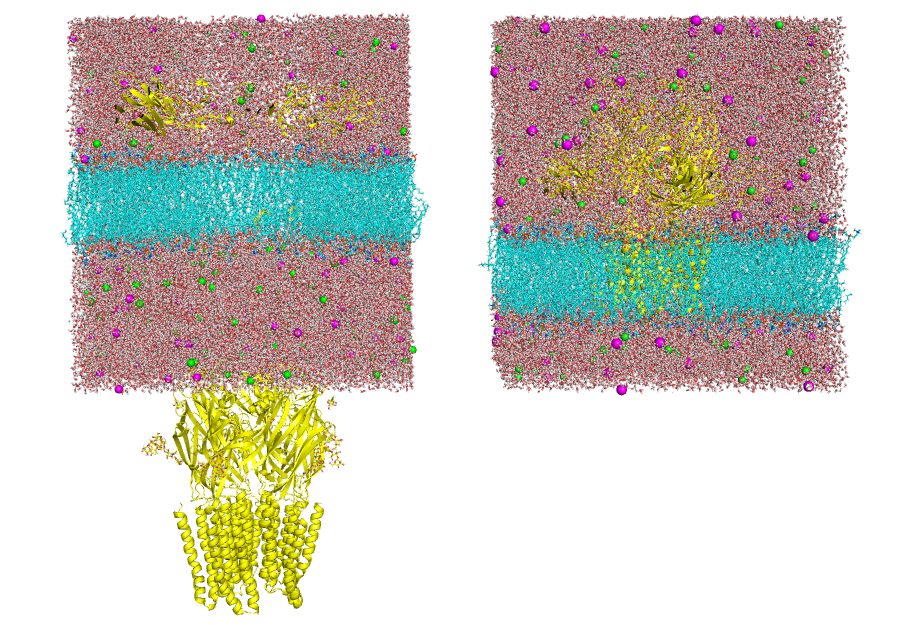
Figure 1. Vizualization of two different input structures files. Left: "Broken" structure; Right: Centered structure Make sure you have fitted the trajectory
Visualize the trajectory given in the
-ct,-rt, or-ltoptions and make sure the PBC has been removed (as shown in Fig 2, right panel). On the other hand, if the trajectory has not been fitted (as shown in Fig 2, left panel) this could generate inconsistent results.Steps:
-
Generate a group that contains both molecules
gmx make_ndx -n index.ndx >1 | 12 >qAssuming 1 is the receptor and 12 is the ligand. This creates a new group (number 20 in this example)
-
remove the PBC
gmx trjconv -s md.tpr -f md.xtc -o md_noPBC.xtc -pbc mol -center -n -ur compact center: 20 (created group) output: 0 -
remove the rotation and translation with respect to the reference structure (optional)
gmx trjconv -s md.tpr -f md_noPBC.xtc -o md_fit.xtc -n -fit rot+trans fit: 20 (created group) output: 0 -
Visualization
Make sure that the trajectory is consistent (as shown in Fig 2, right panel)
-
If the process is not succesful, consider using other options like
-pbc nojump(as suggested here)
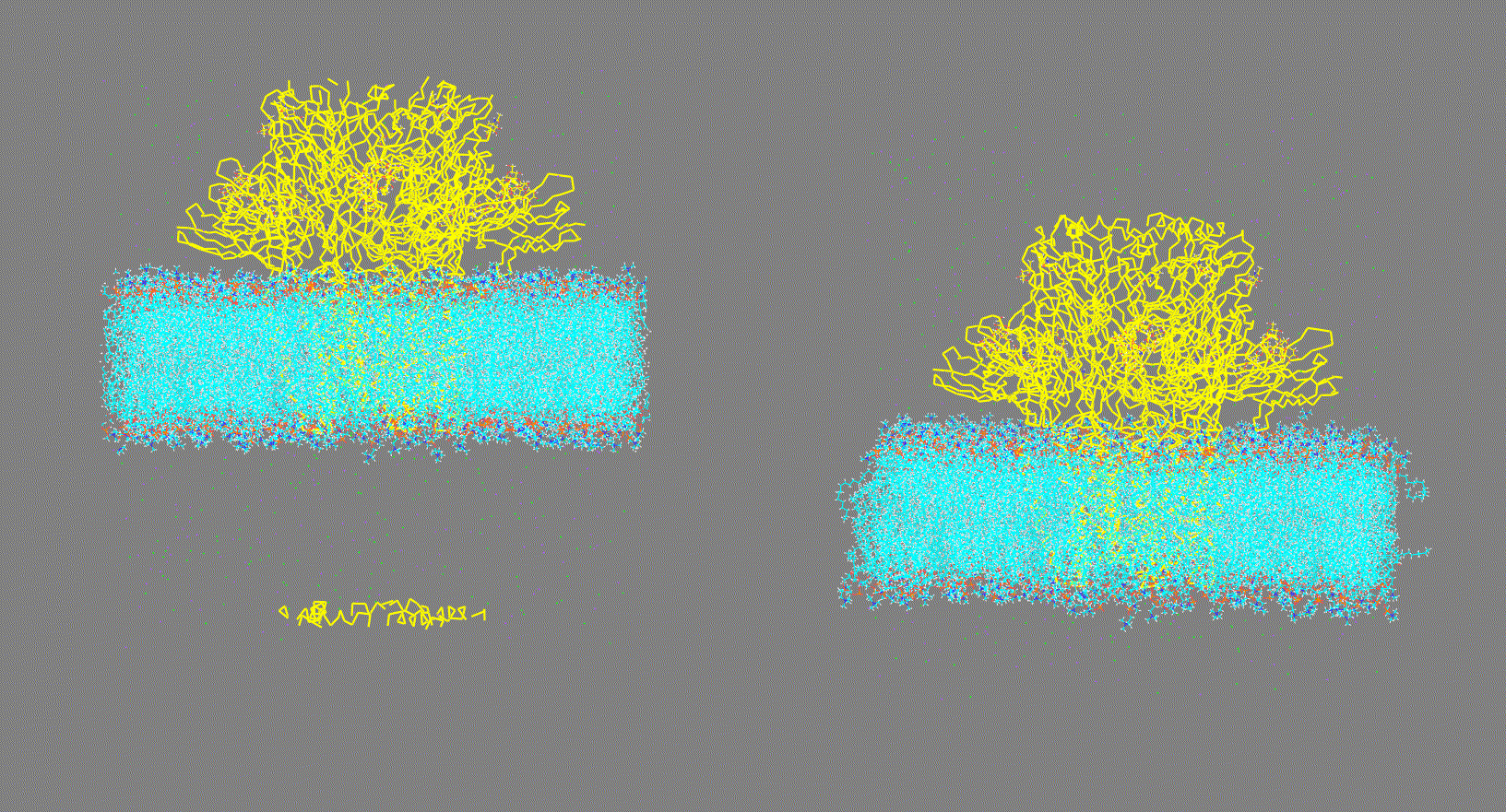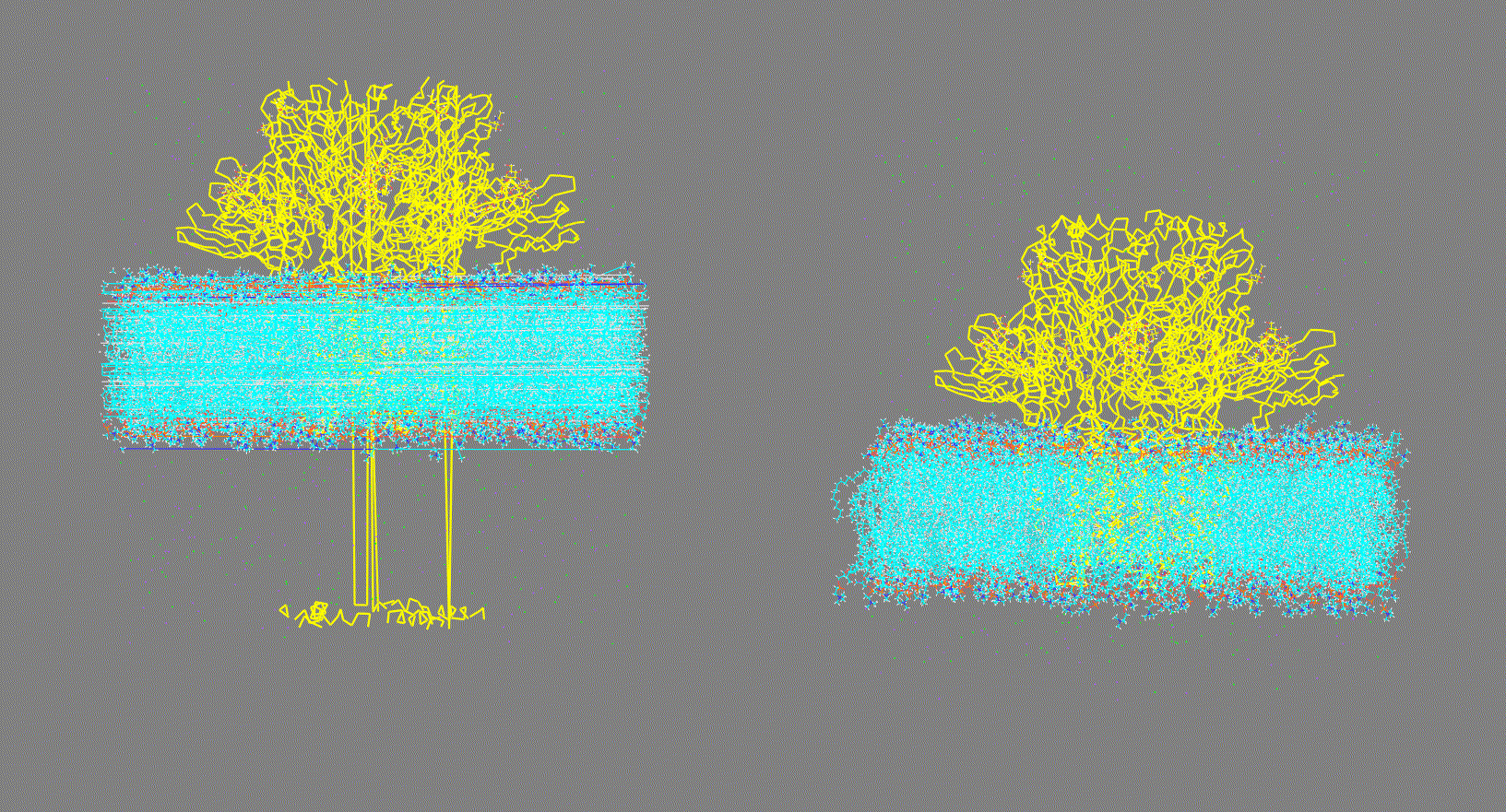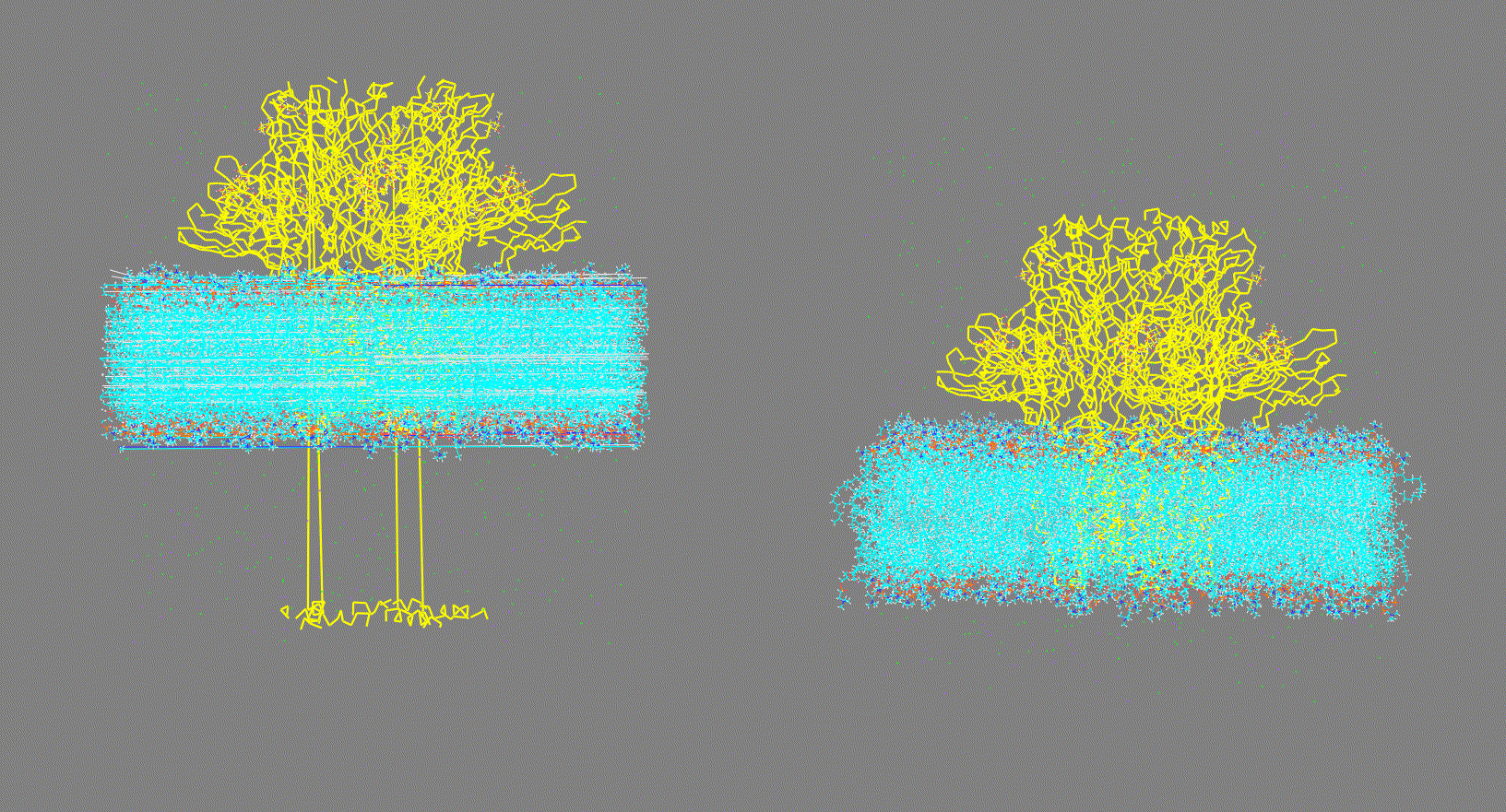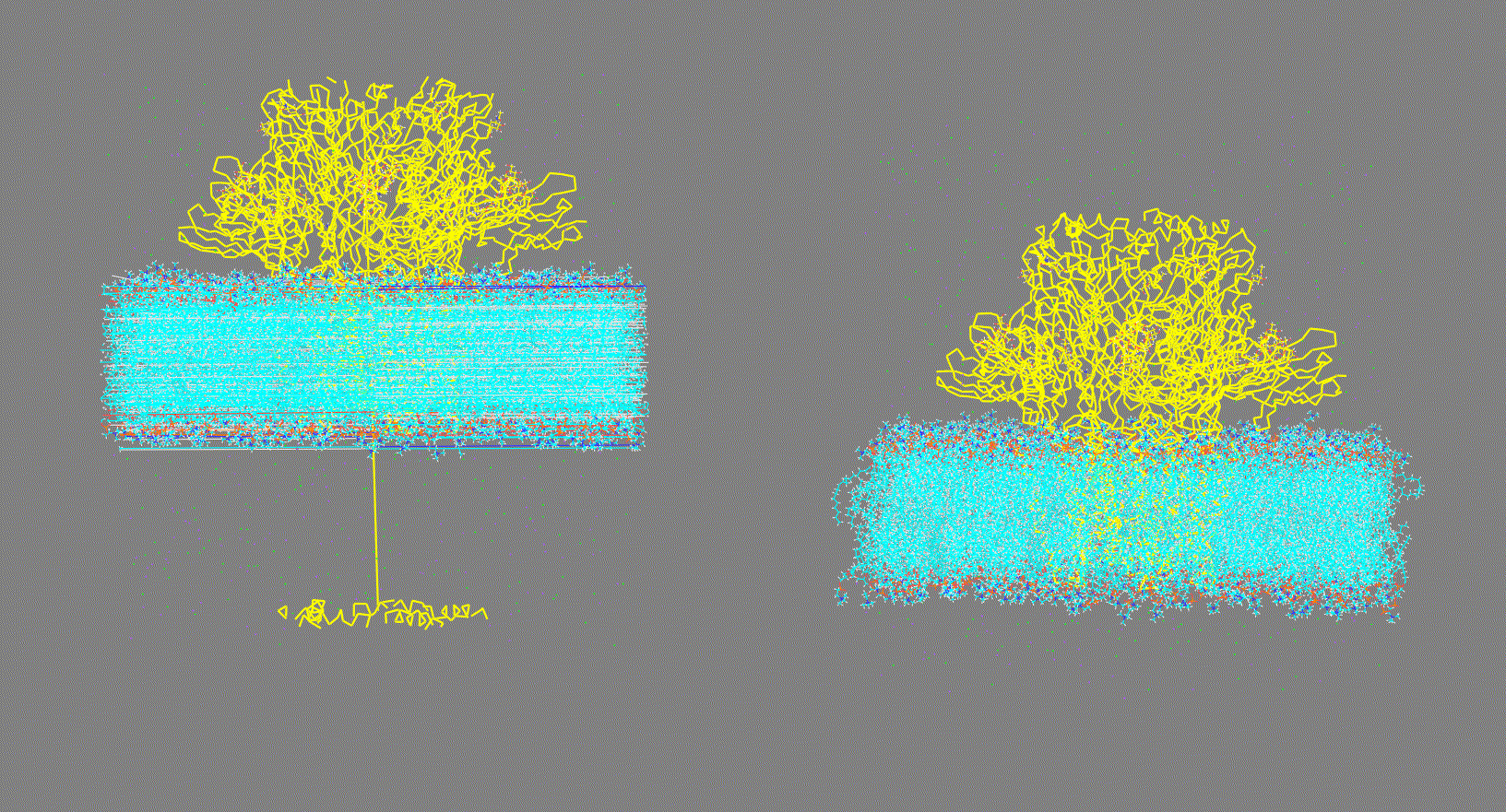
Figure 2. Vizualization of two different input trajectory files. Left: Trajectory with PBC; Right: Trajectory centered, fitted and with PBC removed. -
-
You are trying to calculate the energetic contribution of a very large group. Technically, the energy value should not exceed 7 digits, so if you get a value higher than this, this error will occur. Although
gmx_MMPBSAcan handle very large systems, it cannot determine certain energetic terms. This is asanderlimitation when writing the output file.Possible solutions:¶
- The error could be solved by recompiling
sanderwith some modifications in the output function. However, this is not recommended since the error can be large. Another possible solution could be modifying the parameters of the calculation (solvent model, internal dielectric constant) or just performing the calculation for a part of the system (sub-system).
- The error could be solved by recompiling
-
I get high values for the solvation energy when using PB model
-
When using PB model, inp=2 is used as default to calculate total non-polar solvation free energy, that is, the total non-polar solvation free energy will be modeled as two terms: the cavity term and the dispersion term. The dispersion term is computed with a surface-based integration method closely related to the PCM solvent for quantum chemical programs. Under this framework, the cavity term is still computed as a term linearly proportional to the molecular volume enclosed by SASA.
Possible solutions:¶
-
You may want to try inp=1 and avoid the EDISPER contribution. This way, the total non-polar solvation free energy will be modeled as a single term linearly proportional to the solvent-accessible surface area. Just add
inp=1in the&pbnamelist variables in the input file. See example below: -
Another way to avoid the EDISPER contribution is by modifying the
_GMXMMPBSA_infofile. Changing the value ofINPUT['inp']to 1 and run: -
will avoid the calculation of the EDISPER contribution and report just the ENPOLAR contribution as a term linearly proportional to the molecular volume enclosed by SASA.
Info
Note that these two approximations will yield different values for the non-polar component of the solvation energy (see here). Use one or another depending on your interest.
-
-
Check this publication and see the drawbacks of modeling the total non-polar solvation free energy with two terms, i.e., the cavity term and the dispersion term. Sometimes there are imbalances in the cancellation of error between the two components and this can produce unrealistic non-polar energy values.
NMODE calculation finish in error
The only error reported is probably related to RAM saturation. NMODE calculations require a considerable amount of RAM depending on the number of atoms in your system. The amount of total RAM consumed during the calculation will be: RAM for 1 frame * number of threads
Created: 2023-06-24